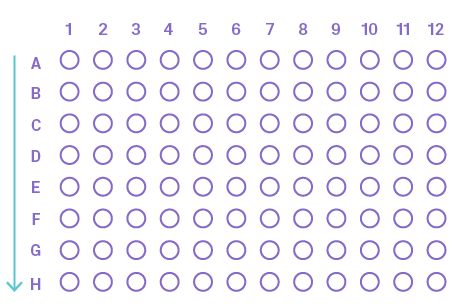GENEWIZ Sanger sequencing protocol

Part 1. Labeling
Two options for doing the outsourcing - non-Qubit based method or Qubit based method.
Guidelines: Samples must be submitted in well-labeled 0.2mL PCR Tubes or in 96-well plates.
It is cheaper per sample when sequencing 96+ reactions.
- $2.95/reaction for 96+ reactions
- $3.45/reaction for 1-95 reactions
Select the California Academy of Sciences Business Reception drop box for pickup. Samples are picked up at 7pm and results are usually available the next afternoon.
Your tube/plate labels need to match your order receipt, so it may be easier to draft your order online and then do your sample preparation.
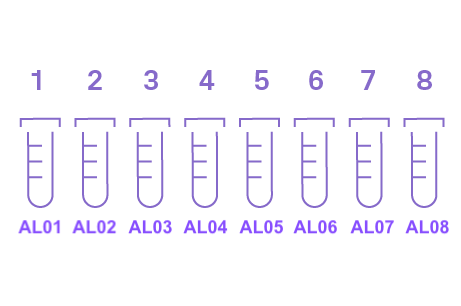
- Tubes must be 8-strip 0.2 mL (200 µL) PCR tubes. You can cut off empty tubes. The order form will generate Sample Numbers and assign Tube IDs. Since they are through Athena Lam’s account, the Tube IDs will always begin with AL.
- Plates: For 48 samples or more, it is recommended to use a 96-well plate for sequencing with 8-strip PCR caps. Plates do not need to be labeled, but be sure to arrange your samples vertically, so Sample 1 is in A1, Sample 2 in B1, etc.
Part 2: Sample Preparation
Option 1: Non-Qubit Method:
- Add 5µL of 5 µM primer to each tube (2.5µL of our standard 10 µM primers)
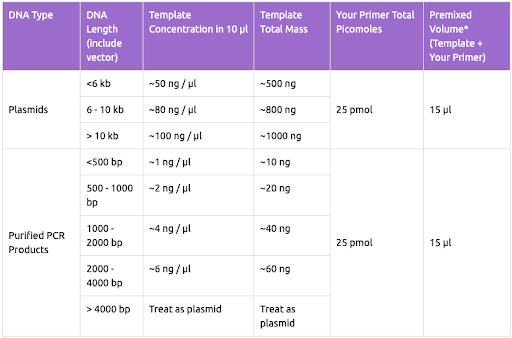
- Add a standard amount of DNA to each tube for your samples (remember to have each direction separate). More DNA is required for longer fragments >1,000 bp.
- Herps - 1µL
- Arachnids - 3µL
- Nudibranchs - 6µL
- Add Millipore water to bring volume in tube up to 15 µL.
Option 2: Qubit Method:
- After ExoSAP-IT, Qubit each sample to determine ng/µL for each sample.
- Calculate amount of DNA to be added to each well to have desired concentration:
- < 500 bp: ~10 ng (H3)
- 500-1,000 bp: ~20 ng (COI, 16S, 28S fragments)
- 1,000-2,000 bp: ~40 ng (18S, 28S full)
- Add 5µL of 5 µM primer to each tube (2.5µL of our standard 10 µM primers).
- Add calculated amount of DNA from Qubit to each well.
- Add Millipore water to bring volume in tube up to 15µL.
Part 3: Setting up sequencing pickup and sample submission
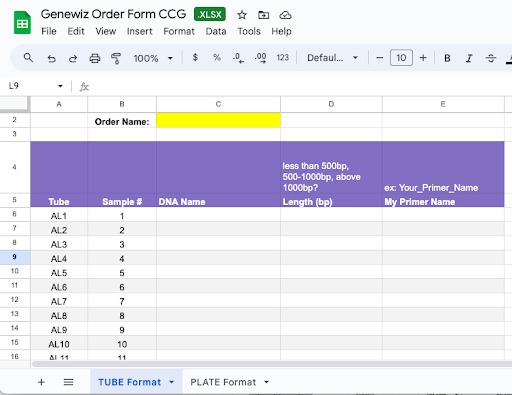
- In the CCG Google Drive, find the Google sheet “Genewiz Order Form CCG”. Make a copy and rename the file to include the Order Name you assign.
- Choose the sheet TUBE Format or PLATE Format according to how you’re submitting your samples.
- Assign your order an Order Name. I suggest including a date.
- Fill in the columns DNA Name and Primer Name according to how you prepared your samples in Part 1.
- Identify the desired Length (bp) range of your PCR product.
- Package your samples for sequencing into a plastic bag and label the bag with your Order Name. Store in 4°C.
- Send this form to Grace at gkim@calacademy.org along with a description of where your samples are located.
- Samples can be submitted for same-day pickup until 5pm.
- Sequences are generally returned the next day. Sequence data is stored online for 2 years, but it is best practice for you to keep your raw sequence files on your personal computer or drive.